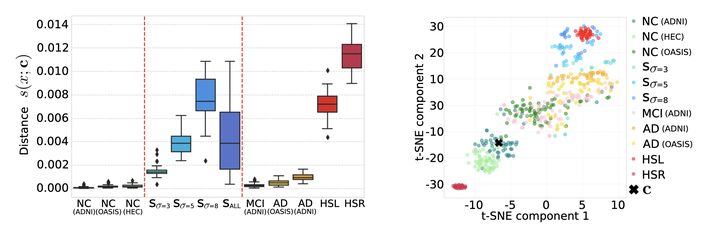
Abstract
Although normal homologous brain structures are approximately symmetrical by definition, they also have shape differences due to e.g. natural ageing. On the other hand, neurodegenerative conditions induce their own changes in this asymmetry, making them more pronounced or altering their location. Identifying when these alterations are due to pathological deterioration is still challenging. Current clinical tools rely either on subjective evaluations, basic volume measurements, or disease-specific deep learning models. This paper introduces a novel method to learn normal asymmetry patterns in homologous brain structures based on anomaly detection and representation learning. Our framework uses a Siamese architecture to map 3D segmentations of the left and right hemispherical sides of a brain structure to a normal asymmetry embedding space, learned using a support vector data description objective. Being trained using healthy samples only, it can quantify deviations-from-normal asymmetry patterns in unseen samples by measuring the distance of their embeddings to the center of the learned normal space. We demonstrate in public and in-house datasets that our method can accurately characterize normal asymmetries and detect pathological alterations due to Alzheimer’s disease and hippocampal sclerosis, even though no diseased cases were accessed for training. Our source code is available at https://github.com/duiliod/DeepNORHA.